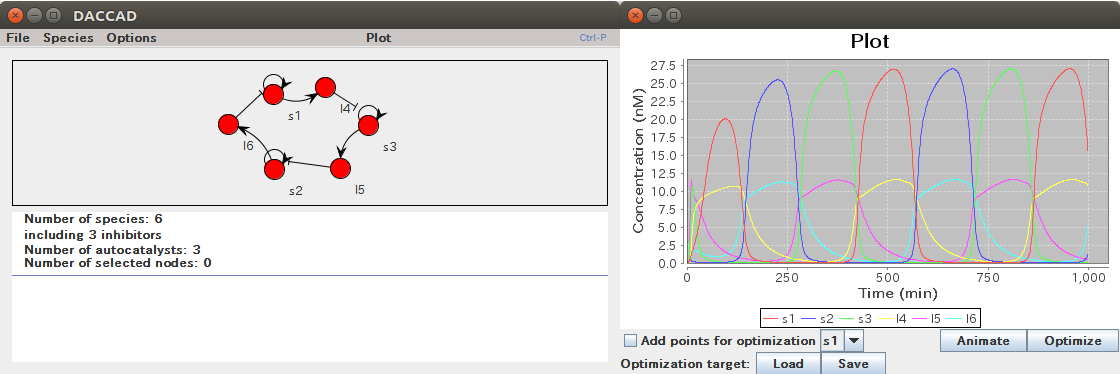
DACCAD (DNA Artificial Circuit Computer Assisted Design)
Latest version download
Contents: the basic executable (DACCAD-1.x.x.jar) and some examples.
You can get the tutorial HERE
What's new in version 1.0
- Color coding for animations: nodes' color represent their dissociation constants, going from blue (low
dissociation constant) to red (high dissociation constant). Templates' color represent how active they are,
red meaning that all the template is currently used to generate its output, blue that most of the template molecules
are free or inhibited, and grey that less than 10
#37; of the template concentration is active. - Now possible to remove individual templates from the data panel.
- Possible to optimize enzyme activities.
- Possible to save optimization profiles.
- Possibility to add comments in .graph files.
- Many bug fixes. Thank you all for taking the time to report them!
Features
- Graph-based design of DNA toolbox systems. Nodes represent signal sequences and arrows DNA templates.
- Ease of use: all parameters have realistic preset values, so that little to no prior knowledge of DNA chemistry is required.
- Complex model simulation, taking into account enzymatic saturation, leaks, and more.
- Local optimization of biochemical parameters using CMA-ES.
- Graph animation, giving a visual interpretation of the behavior.
- System export in SBML or as raw data. A limited version of the model can also be exported in Mathematica.
Notable commands and shortcuts
- Left click on a node without previous selection: select node
- Left click on a node with another node selected: create template from the selected node to the clicked node (can be the selected node itself).
- Left click on the back ground: clear selection.
- Ctrl+a: new sequence.
- Ctrl+z: undo.
- Shift+left click: add node to current selection
Design by TEMPLATED.