Hello
My name is Nathanaël Aubert-Kato. I am currently a lecturer at the department of Information Sciences, from the Ochanomizu University (Tokyo, Japan).
My research focus is on modeling, simulating, and designing molecular programming systems. My long term research goal is to provide methods to develop and program DNA computing-based smart materials.
I follow two research directions:
Bottom-up: considering a specific set of interactions among molecules, create the right model and simulation tools, leading to the discovery of emergent properties
Approach: mass-action kinetics; ODE/PDE; data analysis
Top-down: considering a target abstract behavior, define modules or lower-level behaviors that are required and can be implemented more easily.
Approach: agent-based modeling; evolutionary strategies
contact: naubertkato at is dot ocha dot ac dot jp
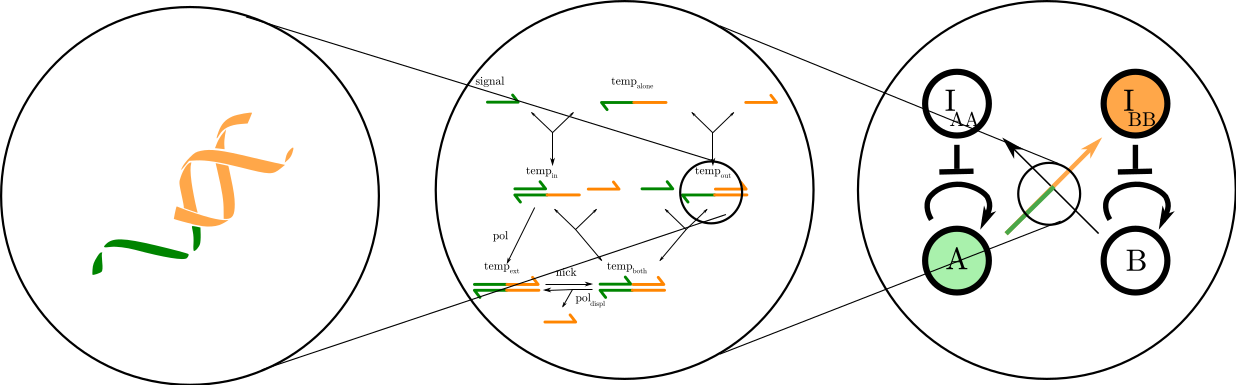
Computing with molecules
Molecular programming is a computing paradigm that relies on chemical reactions among molecules to encode operations. DNA is an excellent material as reactions are strongly dependent on sequence through Watson-Crick base-pairing (A pairs with T, C with G, only complementary sequences can interact). This gives us the possibility to abstract and simplify behaviors to focus on high-level mechanics (see above Figure). In recent years, the field has flourished, with both theoretical proofs of Turing equivalence [Seelig et al., 2006] and real-life, in-vitro implementations (see for instance the review from [Padirac et al., 2013]).
Upcoming conferences/events
-
GECCO2023, Lisbone, Portugal
The Genetic and Evolutionary Computation Conference
-
ALIFE2023, Sapporo, Japan
The 2023 Conference on Artificial Life
-
DNA 29, Sendai, Japan
The 29th International Conference on DNA Computing and Molecular Programming